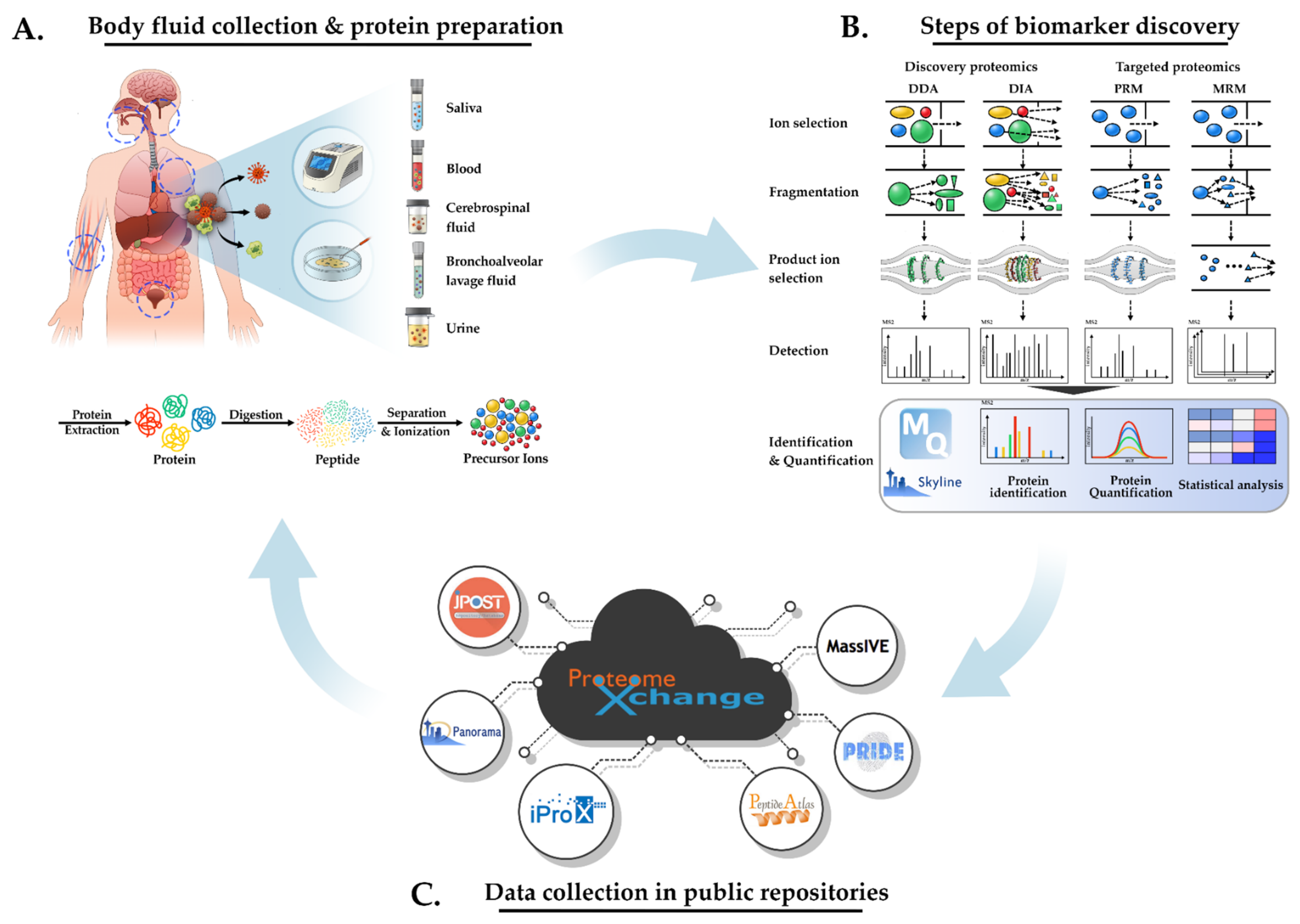
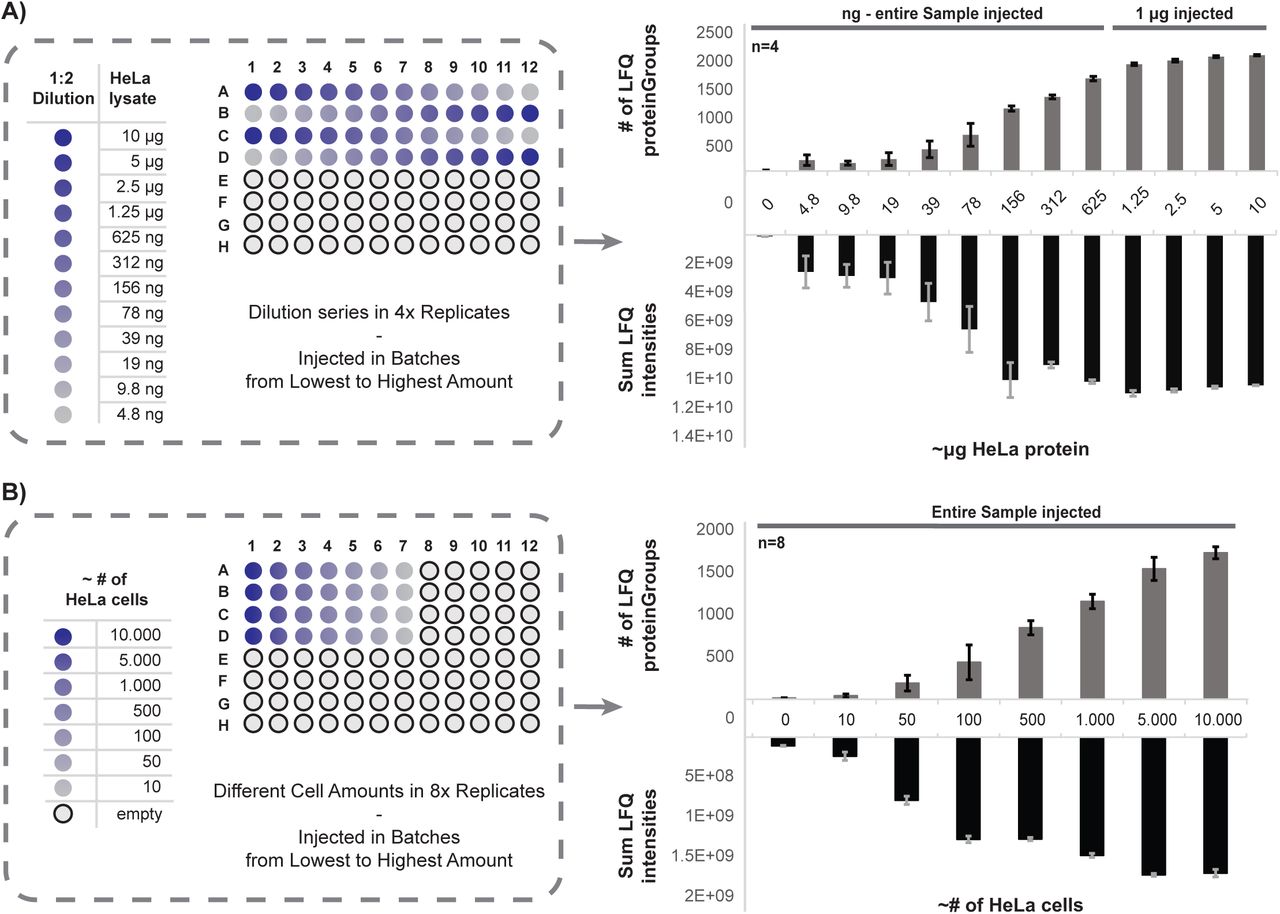
Proteomics for Low Cell Numbers: How to Optimize the Sample Preparation Workflow for Mass Spectrometry Analysis
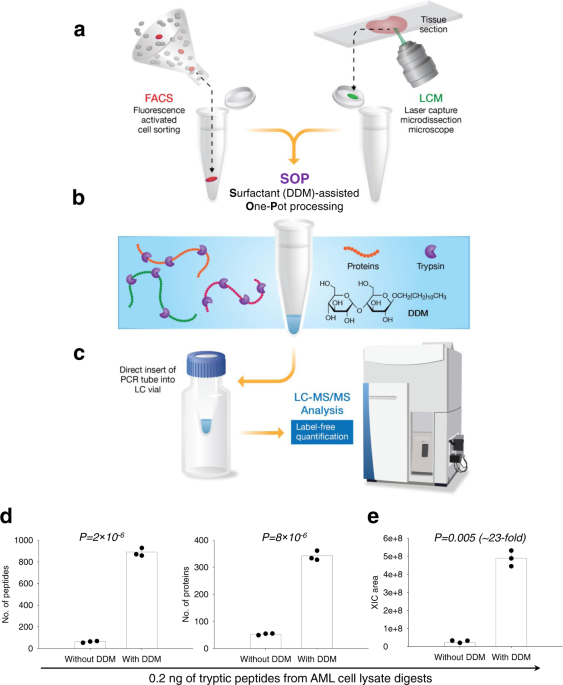
Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics | Communications Biology

SP3-Enabled Rapid and High Coverage Chemoproteomic Identification of Cell-State–Dependent Redox-Sensitive Cysteines - Molecular & Cellular Proteomics

Automated 16-Plex Plasma Proteomics with Real-Time Search and Ion Mobility Mass Spectrometry Enables Large-Scale Profiling in Naked Mole-Rats and Mice | Journal of Proteome Research
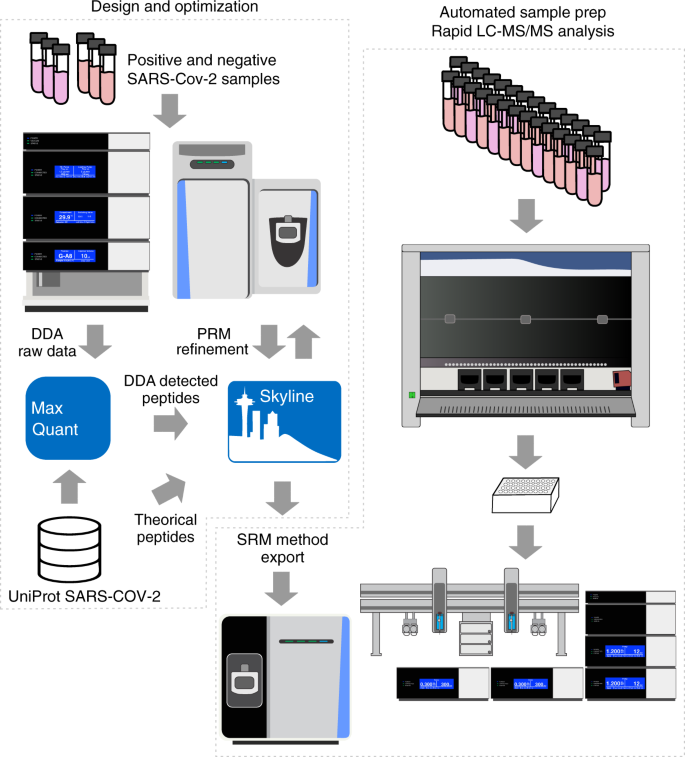
Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts | Nature Communications

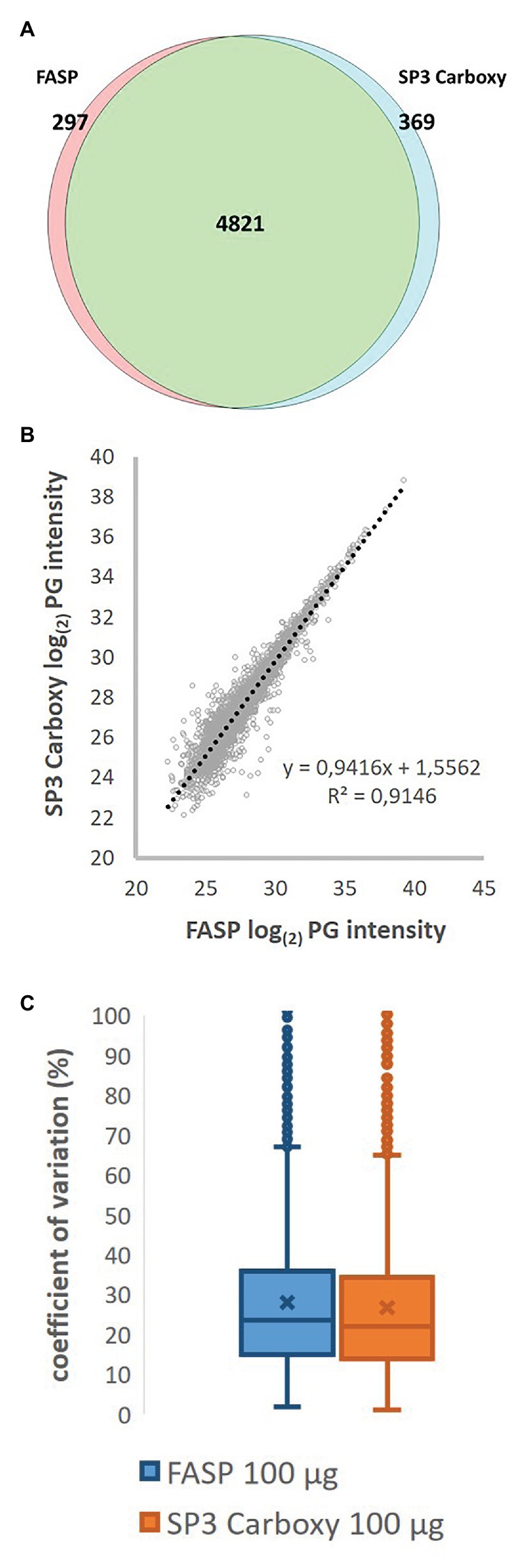
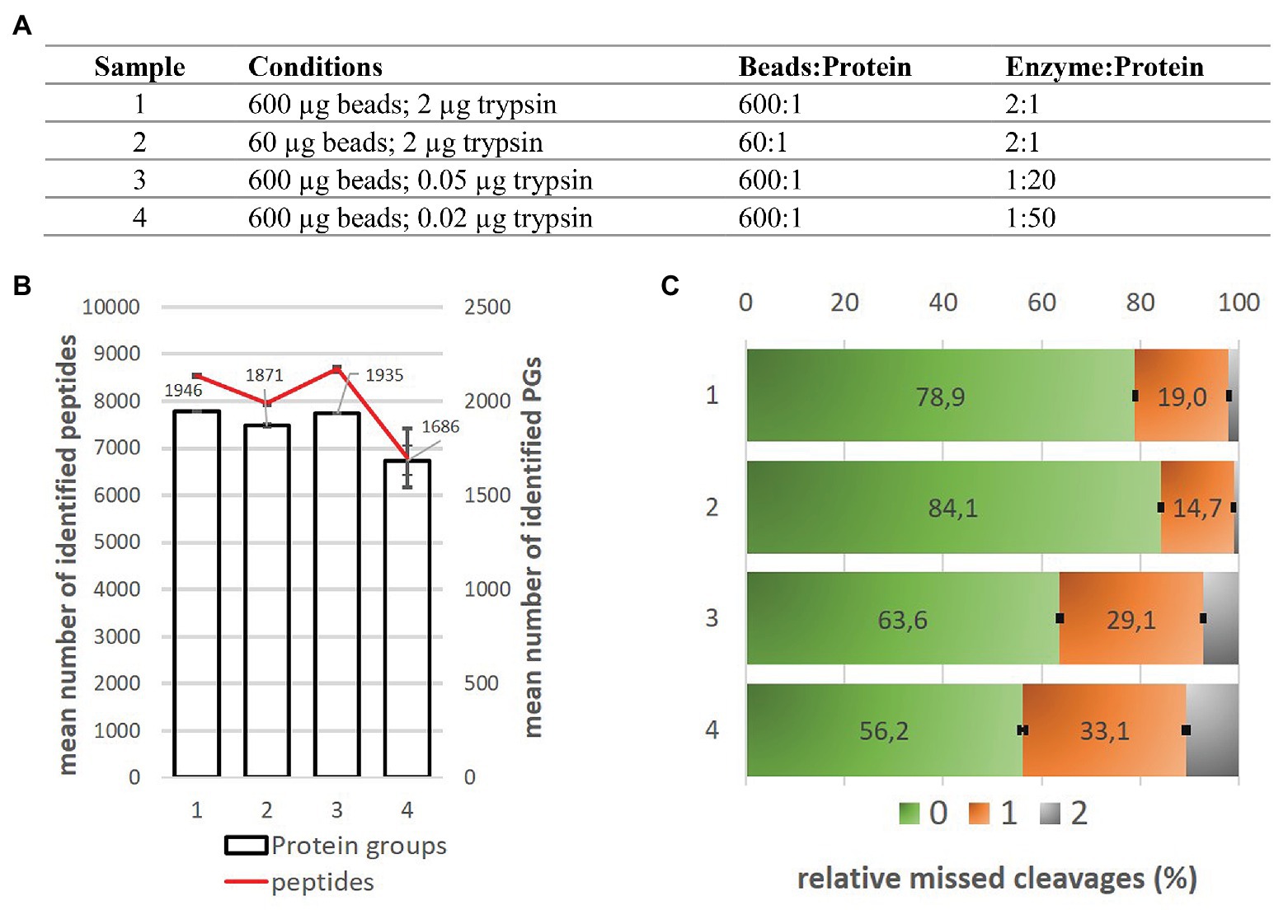
Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics | Journal of Proteome Research

Molecules | Free Full-Text | Mass Spectrometry Advances and Perspectives for the Characterization of Emerging Adoptive Cell Therapies | HTML
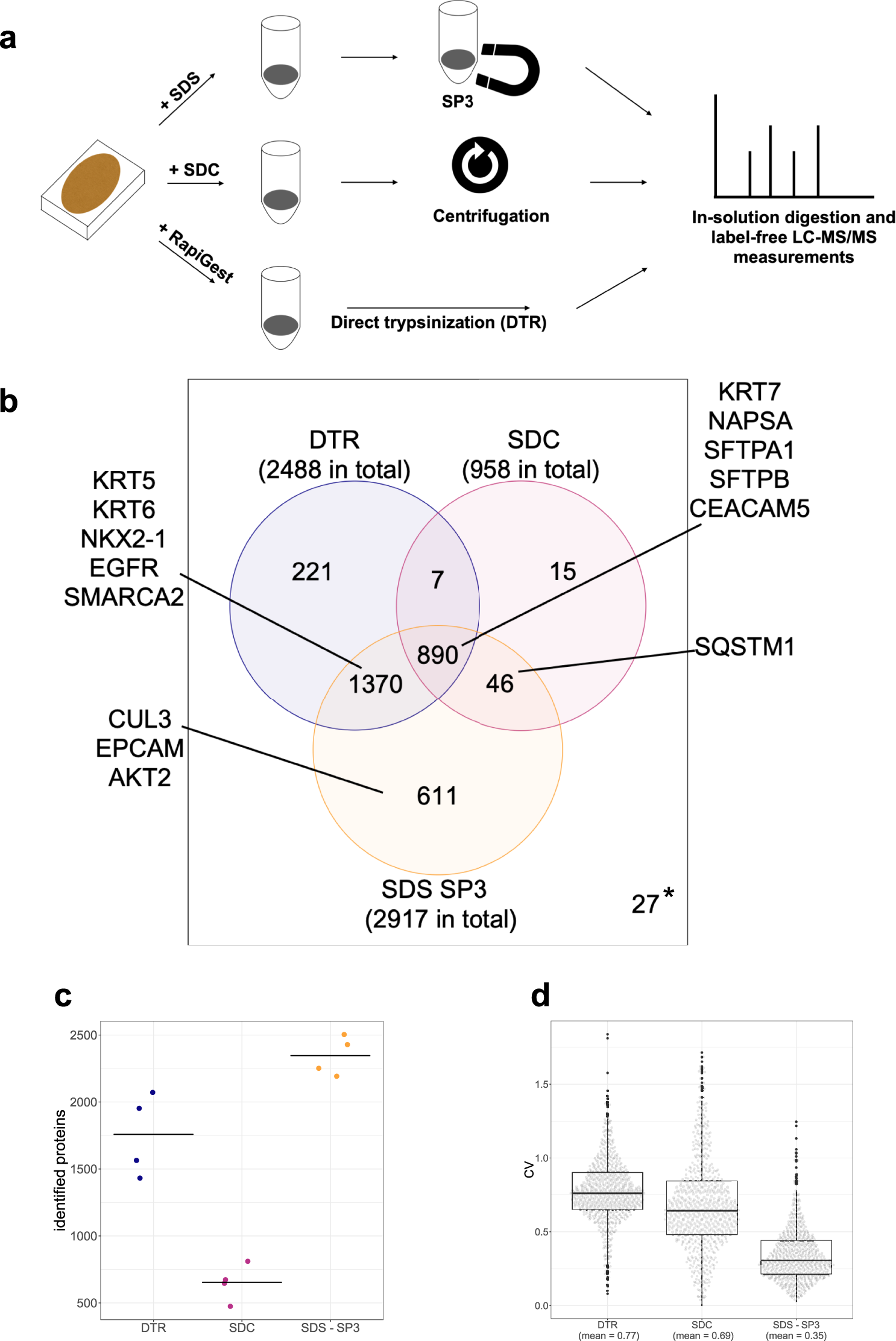
Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories | Nature Communications
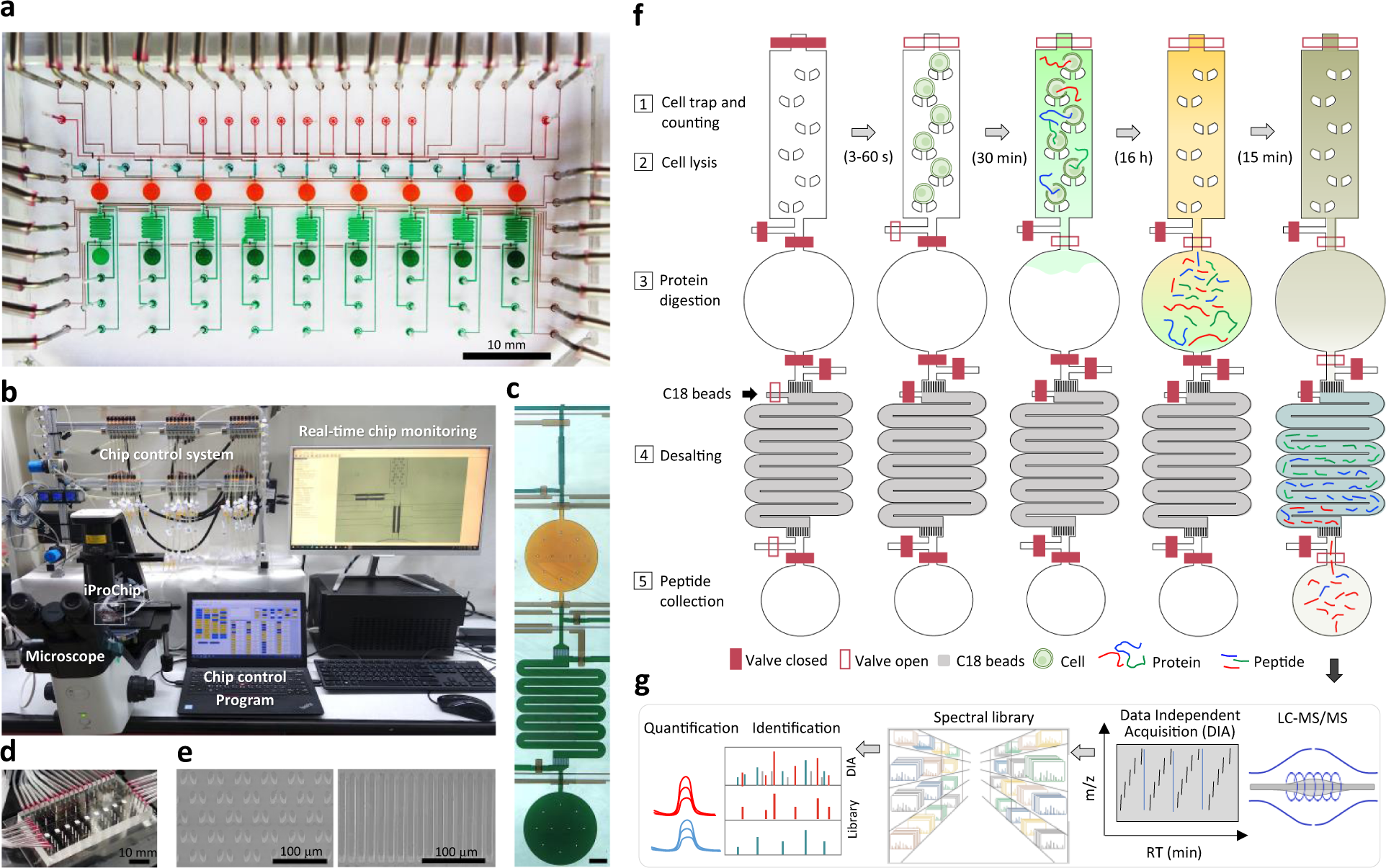
Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry | Nature Communications